Research cluster: Essigsäurebakterien / Subproject |
| Subproject 4B: Detailed Quantitative Metabolome and Fluxome Analysis of Acetic Acid Bacteria with Carbon Labeling Experiments |
| Project time: 01.10.09-31.07.13 |
| Project leader: |
| Dr. Katharina Nöh, Forschungszentrum Jülich GmbH |
| Dr. Marco Oldiges, Forschungszentrum Jülich GmbH |
| Prof. Dr. Wolfgang Wiechert, Forschungszentrum Jülich |
| Acetic acid bacteria like Gluconobacter oxydans are known to oxidize different carbon sources regioselectively in order to produce vitamin C, 2- and 5-ketogluconic acids, dihydroxyacetone or the antidiabetic drug Miglitol. Acetic acid bacteria are valuable and versatile biocatalysts for industrial biotechnology. To deeply understand the cells' metabolism is indispensable when aiming at an increased productivity under industrially relevant conditions. A wide range of omics-tools comprising genomic and metagenomic sequencing, transcriptome, proteome, metabolome, and fluxome profiling are nowadays available. Due to post-transcriptional regulation and unknown enzymatic activities, cellular processes might only be sufficiently reflected by the entirety of all metabolic flux rates, the fluxome, in combination with the set of small-molecule metabolites, the metabolome, as the physiologically most relevant, complementary description of metabolic activity. While metabolome data can be directly accessed, the intracellular in vivo metabolic fluxes have to be indirectly inferred from measured quantities and biochemical network models with the aid of computational tools. Currently the most reliable method for determining these unobservable reaction rates in a metabolically steady state is metabolic flux analysis with 13C-labeling experiments (13C-MFA, [1]): A defined 13C-labeled substrate is incorporated into the carbon backbone of a wide range of metabolites either through exchange or by synthesis. The distribution of labeled carbon traversing along metabolic pathways generates a characteristic imprint of labeling patterns whose mass signature is observed by liquid chromatography coupled mass spectrometry (LC-MS, [2]). |
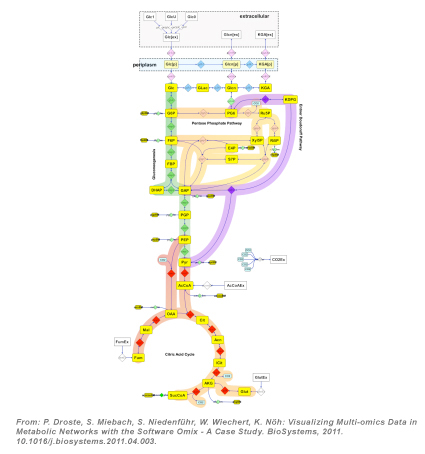 |
As part of an integrative data evaluation approach, 13C-MFA facilitates direct and comprehensive insights into the active metabolic network of acetic acid bacteria. In particular, functional understanding of membrane-bound enzymes in these bacteria would pave the way for more efficient biotransformations and the development of novel oxidative fermentation processes. Metabolome-based 13C-MFA will be used to support the following three key issues of the cluster project:
In our project we conduct carbon labeling experiments in order to characterize wild type and engineered strains of acetic acid bacteria on metabolome and fluxome level. Analyzed with tandem MS analytic devices, data are subsequently used for 13C-MFA modeling purposes. Metabolome-based metabolic flux analyses are conducted in order to assist the hypothesis-driven process of rational strain design with powerful model-based tools. Questions of rational acetic acid bacteria strain optimization as well as of the design of optimal biotechnological processes are addressed. |
 |
References: |
| [3] P. Droste, S. Miebach, S. Niedenführ, W. Wiechert, K. Nöh: Visualizing Multi-omics Data in Metabolic Networks with the Software Omix - A Case Study. BioSystems, 2011. 105(2): p. 154-161. |
| [2] M. Oldiges, S. Lütz, S. Pflug, K. Schroer, N. Stein, and C. Wiendahl: Metabolomics: Current State and Evolving Methodologies and Tools. Appl Microbiol Biotechnol, 2007. 76(3): p. 495-511. |
| [1] W. Wiechert: 13C Metabolic Flux Analysis. Metab Eng, 2001. 3(3): p. 195-206. |
Funded by


New Brochure: Facetten der Genomforschung
Wissenschaftler entschlüsseln die Baupläne des Lebens (german; pdf; 12,3 Mb)
Wissenschaftler entschlüsseln die Baupläne des Lebens (german; pdf; 12,3 Mb)

